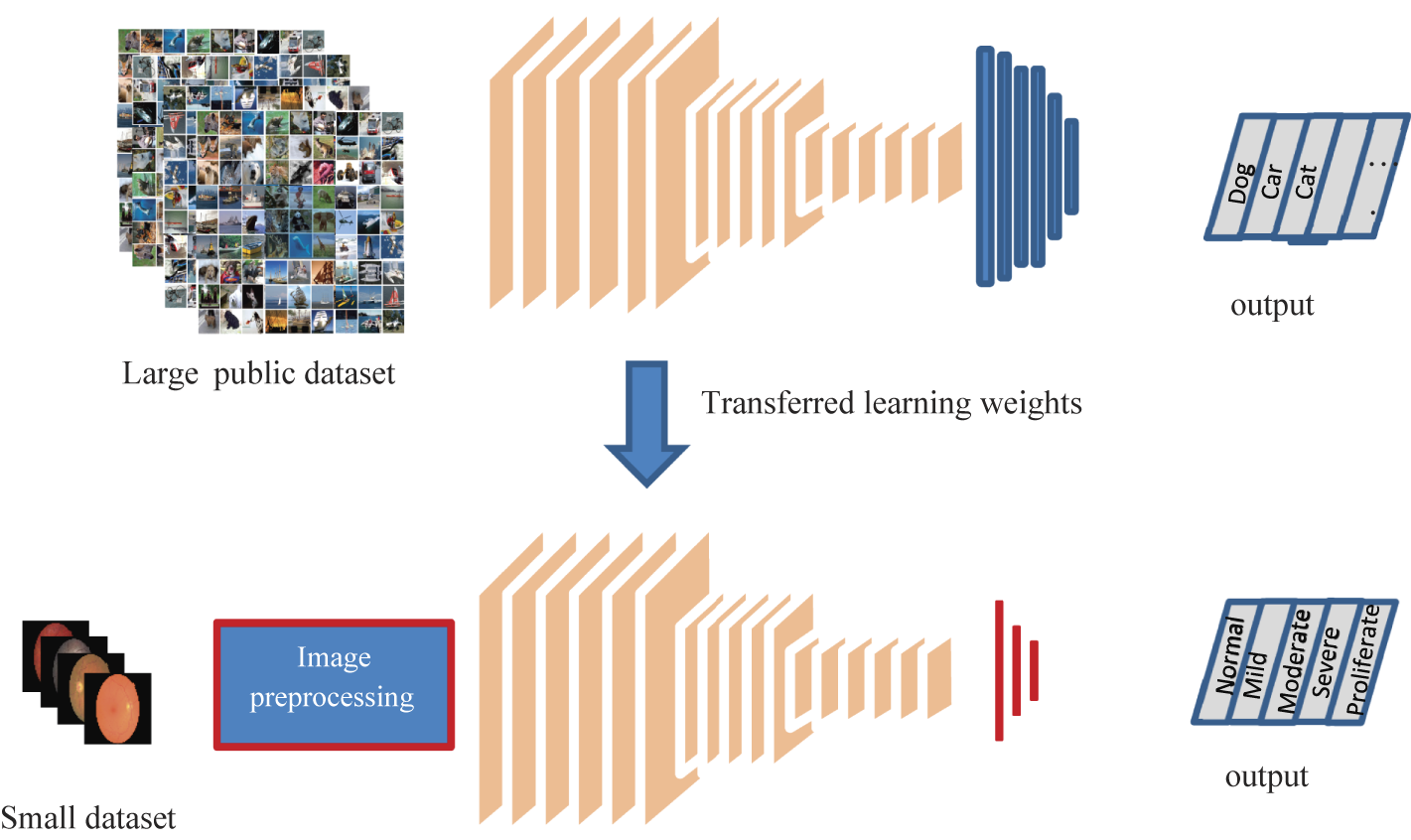
Transfer learning is a machine learning technique that involves leveraging a pre-trained neural network to solve a new problem. The pre-trained neural network is typically trained on a large dataset and has learned to identify and extract meaningful features from the data. By using a pre-trained neural network, we can take advantage of the knowledge that the network has already learned and use it to improve the accuracy of our new model.
In the case of brain tumor detection, we can use a pre-trained neural network like NasNetLarge to improve the accuracy of our model. NasNetLarge is a deep convolutional neural network architecture that has been trained on the ImageNet dataset, which contains millions of images belonging to thousands of classes. It has been shown to perform very well on a variety of image classification tasks.
To use NasNetLarge for brain tumor detection, we can follow these steps:
- Load the pre-trained NasNetLarge model
- Replace the final layer of the model with a new fully connected layer that has a single output node for binary classification (tumor vs. no tumor)
- Freeze the weights of all the layers in the pre-trained model except for the new final layer
- Train the model on our brain tumor dataset, fine-tuning the weights of the new final layer to optimize performance
- Evaluate the performance of the model on a test set
By using transfer learning in this way, we can achieve high accuracy with relatively little data, as we are leveraging the knowledge already learned by the pre-trained model. This can be especially useful in medical imaging applications where obtaining large amounts of labeled data can be challenging.
Here's a Python code for brain tumor detection using the NasNetLarge pre-trained network for transfer learning:
import tensorflow as tf from tensorflow.keras.preprocessing.image import ImageDataGenerator from tensorflow.keras.layers import Dense, GlobalAveragePooling2D from tensorflow.keras.models import Model from tensorflow.keras.optimizers import Adam # Load the NasNetLarge model and its pre-trained weights base_model = tf.keras.applications.NasNetLarge(weights='imagenet', include_top=False) # Freeze the layers of the base model so they're not trained during transfer learning for layer in base_model.layers: layer.trainable = False # Define the top layers of the model for classification x = base_model.output x = GlobalAveragePooling2D()(x) x = Dense(512, activation='relu')(x) predictions = Dense(1, activation='sigmoid')(x) # Define the complete model model = Model(inputs=base_model.input, outputs=predictions) # Compile the model with an Adam optimizer and binary cross-entropy loss model.compile(optimizer=Adam(learning_rate=0.001), loss='binary_crossentropy', metrics=['accuracy']) # Define data generators for training and validation data train_datagen = ImageDataGenerator(rescale=1./255, shear_range=0.2, zoom_range=0.2, horizontal_flip=True) val_datagen = ImageDataGenerator(rescale=1./255) train_generator = train_datagen.flow_from_directory('path/to/training/data', target_size=(224, 224), batch_size=32, class_mode='binary') val_generator = val_datagen.flow_from_directory('path/to/validation/data', target_size=(224, 224), batch_size=32, class_mode='binary') # Train the model with the generators and a specified number of epochs model.fit(train_generator, steps_per_epoch=len(train_generator), epochs=10, validation_data=val_generator, validation_steps=len(val_generator)) # Save the trained model for later use model.save('path/to/saved/model')






No comments:
Post a Comment